Rolling Circle DNA Replication
Circular DNA replication
Gentaur Circular DNA Replication Europe Supply is available at the best prices. Our products are of very high quality, validated by competent laboratories for exclusive use in research. You can order online or send your order to your local contact. If you are not sure of the product choice required for your research, you can always request a selection audit.
DNA replication in bacteria
DNA replication has been well studied in bacteria mainly due to the small size of the genome and the available mutants. E. coli has 4.6 million base pairs (Mbp) on a single circular chromosome and it all replicates in about 42 minutes, starting from a single origin of replication and going around the circle bidirectionally (i.e. in both directions). This means that approximately 1,000 nucleotides are added per second. The process is quite fast and occurs with few errors.
One of the key players is the enzyme DNA polymerase, also known as DNA pol. In bacteria, three main types of DNA polymerases are known: DNA pol I, DNA pol II, and DNA pol III. DNA pol III is now known to be the enzyme required for DNA synthesis; DNA pol I and DNA pol II are primarily required for repair. DNA pol III adds deoxyribonucleotides, each of which is complementary to a nucleotide on the template strand, one by one to the 3′-OH group of the growing DNA strand.
The addition of these nucleotides requires energy. This energy is present in the bonds of three phosphate groups attached to each nucleotide (a nucleotide triphosphate), similar to how energy is stored in the phosphate bonds of adenosine triphosphate (ATP). When the bond between the phosphates is broken and diphosphate is released, the energy released allows for the formation of a covalent phosphodiester bond by dehydration synthesis between the incoming nucleotide and the free 3′-OH group on the growing DNA strand.
Initiation
Replication initiation occurs at a specific nucleotide sequence called the replication origin, where various proteins come together to begin the replication process. E. coli has a single origin of replication (as do most prokaryotes), called oriC, on its single chromosome. The origin of replication is approximately 245 base pairs long and is rich in adenine-thymine (AT) sequences.
Some of the proteins that bind to the origin of replication are important in making single-stranded regions of DNA accessible for replication. Chromosomal DNA is usually wrapped around histones (in eukaryotes and archaea) or histone-like proteins (in bacteria), and is supercoiled, or tightly coiled and twisted on itself. This packaging makes the information in the DNA molecule inaccessible. However, enzymes called topoisomerases change the shape and supercoiling of the chromosome. For bacterial DNA replication to begin, topoisomerase II, also called DNA gyrase, relaxes the supercoiled chromosome.
An enzyme called helicase then separates the DNA strands by breaking the hydrogen bonds between the nitrogenous base pairs. Remember that AT sequences have fewer hydrogen bonds and therefore have weaker interactions than guanine-cytosine (GC) sequences. These enzymes require ATP hydrolysis. As the DNA opens up, Y-shaped structures called replication forks are formed. Two replication forks form at the origin of replication, allowing bidirectional replication and formation of a bubble-like structure when viewed with a transmission electron microscope; as a result, this structure is called a replication bubble. The DNA near each replication fork is coated with single-stranded binding proteins to prevent the single-stranded DNA from rewinding into a double helix.
Once the single-stranded DNA is accessible at the origin of replication, DNA replication can begin. However, DNA pol III can only add nucleotides in the 5′ to 3′ direction (a new DNA strand can only extend in this direction). This is because DNA polymerase requires a free 3′-OH group to which it can add nucleotides by forming a covalent phosphodiester bond between the 3′-OH end and the 5′ phosphate of the next nucleotide. This also means that you cannot add nucleotides if a free 3′-OH group is not available, as is the case with a single strand of DNA.
The problem is solved with the help of an RNA sequence that provides the free 3′-OH end. Because this sequence allows the initiation of DNA synthesis, it is appropriately called a primer. The primer is five to 10 nucleotides long and is complementary to the original or template DNA. It is synthesized by RNAprimase, which is an RNA polymerase. Unlike DNA polymerases, RNA polymerases do not need a free 3′-OH group to synthesize an RNA molecule. Now that the primer provides the free 3′-OH group, DNA polymerase III can now extend this RNA primer, adding DNA nucleotides one by one that is complementary to the template strand.
Elongation
During elongation in DNA replication, nucleotide addition occurs at its maximum rate of about 1000 nucleotides per second. DNA polymerase III can only extend in the 5′ to 3′ direction, which poses a problem at the replication fork. The DNA double helix is antiparallel; that is, one strand is oriented in the 5′ to 3′ direction and the other is oriented in the 3′ to 5′ direction. During replication, a strand, which is complementary to the 3′ to 5′ parent DNA strand, is continuously synthesized towards the replication fork because the polymerase can add nucleotides in this direction. This continuously synthesized strand is known as the leading strand.
The other strand, complementary to the parent DNA from 5′ to 3′, grows away from the replication fork, so the polymerase must move toward the replication fork to start adding bases to a new primer, again in the opposite direction to the replication fork. . It does so until it collides with the previously synthesized strand and then backs off again. These steps produce small pieces of DNA sequence known as Okazaki fragments, each separated by an RNA primer. The Okazaki fragments are named after the Japanese research team and married couple Reiji and Tsuneko Okazaki, who first discovered them in 1966. The strand with the Okazaki fragments is known as the lagging strand and its synthesis is said to be discontinuous.
The leading strand can extend from a single primer, while the lagging strand needs a new primer for each of the short Okazaki fragments. The general direction of the lagging strand will be from 3′ to 5′ and that of the leading strand from 5′ to 3′. A protein called a sliding clamp holds the DNA polymerase in place as it continues to add nucleotides. The sliding clamp is a ring-shaped protein that binds to DNA and holds the polymerase in place. Beyond its role in the initiation, topoisomerase also prevents the DNA double helix from overcooling before the replication fork as the DNA opens; it does this by causing temporary nicks in the DNA helix and then resealing it.
As synthesis proceeds, the RNA primers are replaced by DNA. The primers are removed by the exonuclease activity of DNA polymerase I, and the gaps are filled. The nicks that remain between the newly synthesized DNA (which replaced the RNA primer) and the previously synthesized DNA are sealed by the enzyme DNA ligase which catalyzes the formation of a covalent phosphodiester bond between the 3′-OH end of a DNA fragment. and the 5′ phosphate end of the other fragment, stabilizing the sugar-phosphate backbone of the DNA molecule.
Termination
Once the entire chromosome has replicated, termination of DNA replication must occur. Although much is known about the initiation of replication, less is known about the termination process. After replication, the resulting complete circular genomes of prokaryotes become concatenated, which means that the circular DNA chromosomes are intertwined and must be separated from each other.
This is achieved through the activity of bacterial topoisomerase IV, which introduces double-strand breaks into DNA molecules, allowing them to separate from each other; the enzyme then reseals the circular chromosomes. Resolution of concatemers is a problem unique to prokaryotic DNA replication due to its circular chromosomes. Because both bacterial DNA gyrase and topoisomerase IV are distinct from their eukaryotic counterparts, these enzymes serve as targets for a class of antimicrobial drugs called quinolones.
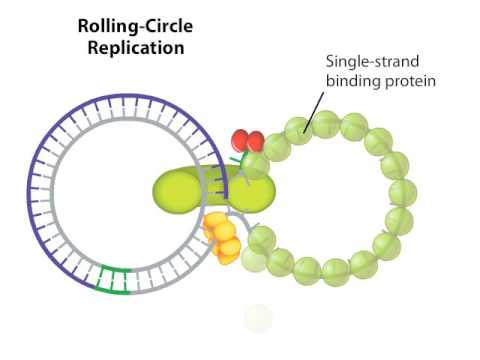
Rolling Circle Replica
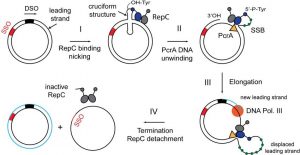
While many bacterial plasmids replicate by a process similar to that used to copy the bacterial chromosome, other plasmids, several bacteriophages, and some eukaryotic viruses use rolling-circle replication. The circular nature of plasmids and the circularization of some viral genomes on infection make this possible. Rolling-circle replication begins with the enzymatic cleavage of one strand of the circular double-stranded molecule at the site of double-stranded origin (dso).
In bacteria, DNA polymerase III binds to the 3′-OH group of the nicked strand and begins unidirectionally replicating the DNA using the unpicked strand as a template, displacing the nicked strand as it does so. Completion of DNA replication at the original nick site results in the complete displacement of the nicked strand, which can then recirculate into a single-stranded DNA molecule. RNA-primase then synthesizes a primer to initiate DNA replication at the single-stranded site of origin (so) of the single-stranded DNA (ssDNA) molecule, resulting in a double-stranded DNA (dsDNA) molecule identical to the other. circular DNA molecule.